Artificial Intelligence and Machine Learning: Unit II: Probabilistic Reasoning
Probabilistic Reasoning and Bayesian Networks
Probabilistic Reasoning - Artificial Intelligence and Machine Learning
Probability based reasoning is the same as inferring directly from knowledge that can be given a probability rating based on the amount of uncertainty present.
Probabilistic
Reasoning and Bayesian Networks
The
concept:
Probability
based reasoning is the same as inferring directly from knowledge that can be
given a probability rating based on the amount of uncertainty present. The
uncertainties can arise from an inability to predict outcomes due to
unreliable, vague incomplete or inconsistent knowledge. The probability of an
uncertain event is a measure of the degree of likelihood of occurence of that
event.
When
one takes it for granted that certain parameters exist or known with certainty,
solutions can be found out. Belief revision takes place whenever one encounters
a piece of information that decreases the belief of the fact.
The
source for all uncertainties is the real world. The following sources
constitute the major causes of uncertainties -
i)
Most information is not obtained personally but got from sources on which one
has a lot of belief. Uncertainty prevails when the source does not send
information in time or the information provided is not understood.
ii)
In laboratory, experiments one takes it for granted that the equipment is
properly calibrated and error free. If the equipment is faulty, the picture one
gets about the situation is not correct which leads to uncertainty.
iii)
Experimental errors like parallax errors are also sources of uncertainty.
iv)
Imprecision in natural language is also a source of uncertainty.
v) A
random event occurring is a major source of uncertainty.
vi)
In judicial courts one may have seen or heard delivering judgements acquitting
the accused for -
a)
Lack of evidence
b)
Lack of certainty in evidence thereby giving the benefit of doubt to the
accused.
To
handle these uncertain data, probability is the oldest technique available.
Probabilistic reasoning is sometimes used when outcomes are unpredictable. For
example, when a physician examines a patient, the patient's history, symptoms
and test results provide some, but not conclusive evidence of possible
ailments. This knowledge, together with the physician's experience with
previous patients, improves the likelihood of predicting the unknown event, but
there is still much uncertainty in most diagnoses.
One
way to express confidence about such event is through probability. It is a mere
number that expresses the chance of something happening or not happening.
Representing
knowledge in uncertain domain:
Independence
and conditional independence relationships among variables can greatly reduce
the number of probabilities that need to be specified in order to define the
full joint distribution. There is technique called as Bayesian Network which
can be used to represent the dependencies among variables and to give a concise
specification of any full joint probability distribution.
Bayesian Network
1. Definition
It
is a data structure which is a graph, in which each node is annotated with
quantitative probability information.
The
nodes and edges in the graph are specified as follows:-
1) A
set of random variables makes up the nodes of the network. Variables may be
discrete or continuous.
2) A
set of directed links or arrows connects pairs of nodes. If there is an arrow
from node X to node Y, then X is said to be a parent of Y.
3)
Each node Xi, has a conditional probability distribution P(Xi |
Parents(Xi)) that quantifies the effect of the parents on the node.
4)
The graph has no directed cycles (and hence is a directed, acyclic graph, or
DAG).
The
set of nodes and links is called as topology of the network. The topology of
the network specifies the conditional independence relationships that hold in
the domain.
The
intutive meaning of an arrow between two nodes X and Y, in a properly
constructed network, is that "X has direct influence on Y". The
domain expert is a person who can identify such influence associations.
Once
Bayesian network topology is specified then conditional probability
distribution for each variable is specified. For specifying continuous
probability distribution for a variable, its parent information is required.
The
combination of topology and the conditional distributions is sufficient to
specify the full joint distribution for all the variables.
Consider
our example of simple world consisting of variables toothache, cavity, catch
and weather. Weather is surly independent of other variables. Toothache and
Catch are conditionally independent if given the information of cavity.
This
relationships are represented by Bayesian network as shown below:
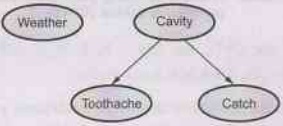
Fig. 7.3.1 A simple Bayesian network in which
Weather is independent of other three variables and Toothache and Catch are
conditionally independent, given the cavity
Note
1)
The conditional independence of toothache and catch given the cavity is
indicated by the absence of a link between toothache and catch.
2)
The network represents the fact that cavity is a direct cause of toothache and
catch.
3)
No direct causal relationship exists between toothache and catch.
Consider
another example of burglar alarm installed at A's home. This alarm notifies in
case of burglary and can occasionally respond to minor earthquakes. There are
two neighbours to A, M and J. M and J promised to call A in office when they
hear alarm. J always calls A when he hears alarm, but many times he is confused
between telephone ring and burglar alarm. M many a times misses alarm because
he keeps on listening to loud music. Given the evidence of who has or has not
called, we are to estimate the probability of burglary.
The
Bayesian network for above problem along with the associated probabilities is
shown below:
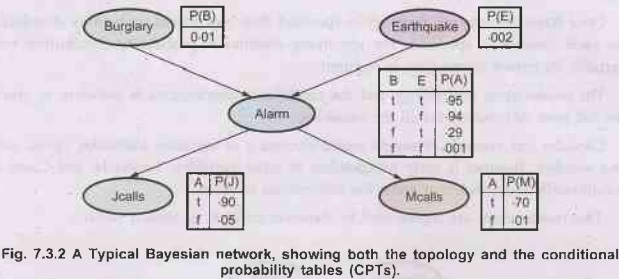
Note In the figure, in the CPTS, the letters B, E, A, J and M stand
for Burglary, Earthquake, Alarm, Johncalls, Marycalls respectively.
1)
In the figure, each distribution is shown as conditional probability table.
Each row in the table contains the conditional probability of each node value
for a 91 conditioning case. A conditioning case is just a possible combination
of values for the parent nodes. Each row must sum upto 1 because the entries represent
an exhaustive set of cases for the variable.
2)
For boolean variable if it is true and its probability is know to be 'P' then
when it is false its probability is 1-P and it is ommited from the table
(because it can be deduced from 'P').
3) A
table for a boolean variable with k boolean parents contains 2k independently
specifiable probabilities. A node with no parents has only one row,
representing the prior probabilities of each possible value of the variable.
2. The Semantics of Bayesian Networks
There
are two ways through which semantic (meaning) of Bayesian network can be
understood.
One
way is to view network as a representation of the joint probability
distribution. This view helps to constructs networks. Second way is to view a
network as an encoding of a collection of conditional independence statements.
This view helps in designing inference procedure semantically both views are
equivalent.
Understanding
semantic of Bayesian network method:
•
Representing the full joint distribution:
1)
Every entry in the full joint probability distribution (hereafter abbreviated
as "joint") can be calculated from the information in the network.
2) A
generic entry in the joint distribution is the probability of a conjunction of
particular assignments to each variable, such as P(X1 = x1 ^.....^xn
= xn)
follg3)
The notation P(x1,...,xn) is used as an abbrevation for
this.
4)
The value of this entry is given by the formula.
P(x1,...,xn) = Пnj=1
P(xi | parents (Xi)) ……(7.3.1)
where
parents (xi) denotes the specific values of the variables in parents
(xi).
5)
Thus, each entry in the joint distribution is represented by the product of the
appropriate elements of the Conditional Probability Tables (CPTs) in the
Bayesian network. The CPTs therefore, provide a decomposed representation of
the joint distribution.
We
can calculate the probability that the alarm has sounded, but neither a
burglary nor an earthquake has occurred and both J and M call. We use single
letter names for the variables:
P(j
Ʌ m Ʌ a Ʌ⌐ b Ʌ⌐ e) = P(j | a) P(m | a) P(a |⌐ b Ʌ⌐ e) P(⌐ b) P(⌐e)
=
0.90 × 0.70 × 0.001 × 0.999 x 0.998 = 0.00062
6)
Remember that the full joint distribution can be used to answer any query about
the domain.
If a
Bayesian network is a representation of the joint distribution, then it too can
be used to answer any query, by summing all the relevant joint entries. A
method for constructing Bayesian network:
1)
We rewrite the joint distribution in terms of a conditional probability, using
the product rule.
P(x1,...,xn)
= P(xn | Xn-1,...,X1) P(xn-1,...,x1).
2)
Then the process is represented, reducing each conjunctive probability to a
conditional probability and a smaller conjunction. We end up with one
bigproduct.
P(x1,...,xn)
= P(xn | xn-1,..., x1)P(xn-1 xn-2
,...,x1).....P(x2 | x1) P(x1)
= IIni=1
P(xi | xj-1,….x1) …..(7.3.2)
3)
Above identity holds true for any set of random variables and is called the
Chain rule. Comparing it with equation (7.3.1). We see that the specification
of the joint distribution is equivalent to the general assertion that, for
every variable Xi in the network,
P(Xi
Xi-1,....X1) = P(Xi | Parents(Xj)),
provided
that parents (Xi) (Xi-1,....X1}. This last
condition is satisfied by labeling the nodes in any order that is consistent
with the partial order implicit in the graph structure.
4)
Bayesian network is correct representation of the domain only if each node is
conditionally independent of its predecessors in the node ordering, given its
parents.
5)
In order to construct a Bayesian network with the correct structure for the
domain, we need to choose parents for each node such that this property holds.
Intuitively, the parents of node X; should contain all those nodes in Xi,....Xi-1
that directly influence Xi.
For
example: Suppose we have completed the network
in Fig. 7.2.2 except for the choice of parents for M calls, M calls is
certainly influenced by whether there is a burglary or an earthquake, but not
directly influenced. Intitutively, our knowledge of the domain tells us that
these events influence M's calling behaviour only through their effect on the
alarm. Also given the state of the alarm, whether J calls has no influence on
M's calling. Formally speaking, we believe that the following conditional
independence statement holds:
P(M
Calls |J Calls, Alarm, Earthquake, Burglary) = P(M calls | Alarm).
•
Compactness and node ordering:
Bayesian
network are compact and they possess a property of being locally structured
(also called as sparse systems). In a locally structured system, each sub
component interacts directly with only a bounded number of other components,
regardless of the total number of components.
⇒ Local
structure is usually associated with linear rather than exponential growth in
complexity.
⇒ We assume
'n' boolean variables for simplicity, then the amount of information on needed
to specify each conditional probability table will be at most 2k numbers. Where
each random variable is influenced by 'k' other variables.
⇒ The
complete network can be specified by n2k numbers.
With
some values of 'n' the joint distribution contains 2n numbers.
⇒ To make
this concrete, suppose we have n = 30 nodes, each with five parents (k=5). Then
the Bayesian network requires 960 numbers, but the full joint distribution
requires over a billion.
Ordering
of nodes in Bayesian network:
1)
The correct order in which to add nodes is to add the "root causes"
first, then the variables they influence, and so on, until we reach the
"leaves", which have no direct causal influence on the other
variables.
2)
If wrong order is choosen we get more complicated network.
For
example: Consider network shown in following diagram.
The
network construction process goes as follows:
i)
Adding M calls: No parent.
ii)
Adding J calls: If M calls, that probably means the alarm has gone off, which
ofcourse would make it more likely that J calls. Therefore J calls needs M
calls as a parent.
iii)
Adding alarm: Clearly, if both call, it is more likely that the alarm has gone
off than if just one or neither call, so we need both M calls and J calls as
parent.
iv)
Adding burglary: If we know the alarm state, then the call from J or M might
give us information about phone ringing or M's music, but not about burglary :
P(Burglary Alarm, J calls, M calls) = P(Burglary | Alarm).
Hence
we need just alarm as parent.
v)
Adding earthquake: If the alarm is on, it is more likely that there has been an
2 earthquake. (The alarm is an earthquake detector of sorts). But if we know
that there has been a burglary, then that explains the alarm, and the
probability of an earthquake would be only slightly above normal. Hence, we
need both Alarm and Burglary as parents. The network is shown below.
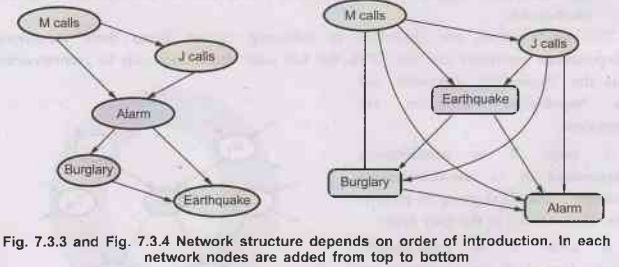
•
Consider example of a very bad node ordering as shown
in the Fig. 7.2.4: M calls, J calls, Earthquake, Burglary, Alarm are nodes.
This network requires 31 distinct probabilities to be specified exactly the
same as the full joint distribution. It is important to realize, however, that
any of the three networks can represent exactly the same joint distribution.
The last two versions simply fail to represent all conditional independence
relationships and hence end up specifying a lot of unnecessary numbers instead.
3. Conditional Independence Relations in Bayesian Networks
One
can start from a "topological" semantics that specifies the
conditional independence relationships encoded by the graph structure, and from
these we can derive the "numerical" semantics. The topological
semantics is given by either of the following specifications, which are
equivalent.
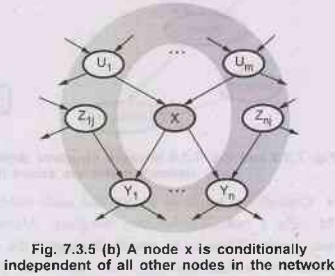
1) A
node is conditionally independent of its non-descendants, given its parents. For
example, in Fig. 7.3.2 J calls is independent of Burglary and Earthquake, given
the value of Alarm.
2) A
node is conditionally independent of all other nodes in the network, given it’s
parents, children, and children’s parents- that is, given its Markov blanket.
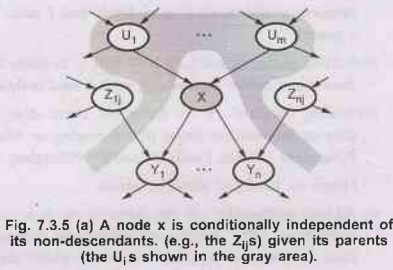
For
example: Burglary is independent of J calls and M calls given Alarm and
Earthquake.
This
specifications are illustrated in following figures. From these conditional
independence assertions and the CPTs, the full joint distribution can be reconstructed;
thus the "numerical" semantics and the "topological"
semantics are equivalent.
A
node X is conditionally independent of its non-descendants (example - - The Z1js)
given its parents(the U; is shown in the gray area).
As
node X is conditionally on independent of all other nodes in the network given its
Markove blanket (the gray area).
Efficient
representation of conditional representation:
If
the maximum number of parents k is smallish, filling in the CPT for a node,
would require up to O(2k) numbers.
To
avoid this big relationship number canconical distribution is used. In this a
complete table is specified by naming the patterns supplied with parameters,
which can describe relationship which are necessary. A deterministic node represents
canonical distribution. A deterministic node is a node whose value is exactly
specified by the value of its parents with no uncertainity.
The
uncertain relationships are often represented by noisy-OR relationships.
[Noisy-OR is generalization of logical OR]. The noisy-OR model allows
uncertainity about the parent to cause the child to be true i.e. the causal
relationship between parent and child would be inhibited.
For
example: A patient could have cold (parent) but not exhibit a fever (child).
For noisy-OR
model it is required that all the possible effects of parent must be listed. It
should be clear that inhibition of one parent is independent of other parent.
The Hybrid Bayesian Network
A
network with both discrete and continuous variables is called as hybrid
Bayesian network. For representing continuous variable its discretization is
done in terms of intervals (because it can have infinite values).
To
specify a hybrid network, we have to specify two new kinds of distributions.
The conditional distribution for a continuous variable given discrete or
continuous parents; and the conditional distribution for a discrete variable
given continuous parents.
Consider
the simple example in following diagram, in which a customer buys some fruit
depending on its cost, which depends in turn on the size of the harvest and
whether the government's subsidy scheme is operating. The variable Cost is
continuous
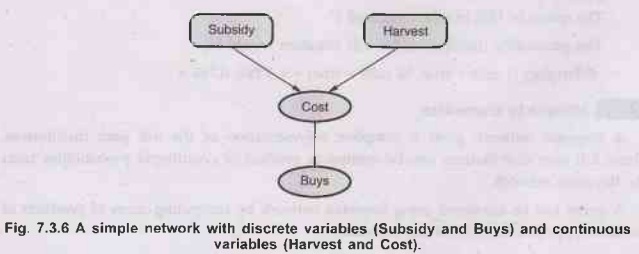
and
has continuous and discrete parents; the variable Buys is discrete and has a
continuous parent.
For
the cost variable, we need to specify P(Cost Harvest, Subsidy). The discrete
parent is handled by explicit enumeration that is, specifying both P(Cost |
Harvest, Subsidy) and P(Cost Harvest, Subsidy). To handle Harvest, we specify how
the distribution over the cost 'c' depends on the continuous value 'h', of
Harvest. In other words, we specify the parameter of the cost distribution as a
function of 'h'.
Inferencing in Bayesian Networks
1. Exact Inference in Bayesian Networks
For
inferencing in probabilistic system, it is required to calculate posterior
probability distribution for a set of query variables, where some observed
events are given. [That is we have some values attached to evidence variables].
•
Notation Revisited:
The
notation used in inferencing is same as the one used in probability theory.
X:
Query variable.
E:
The set of evidence variables E1,....., Em and 'e' is the
perticular observed event. Y: The set of non-evidence variables Y1,
Y2,..... Yk [Non-evidence variables are also called as
hidden variables].
X:
It the complete set of all the types of variables, where X = {X} U E U Y.
•
Generally the query requires the posterior
probability distribution P(X | e) [assuming that query variable is not among
the evidence variables, if it is, then posterior distribution for X simply
gives probability 1 to the observed value]. [Note that query can contain more
than one variable. For study purpose we are assuming single variable].
•
Example: In the burglary case, if the observed event
is Jcalls = true and Mcalls true.
The
query is 'Has burglary occured?'
The
probability distribution for this situation would be,
P(Burglary
| J calls = true, M calls = true) = < 0.284, 0.716 >
2. Inference by Enumeration
A
Bayesian network gives a complete representation of the full joint
distribution. These full joint distributions can be written as product of
conditional probabilities from the Bayesian network.
A
query can be answered using Bayesian network by computing sums of products of
conditional probabilities from the network.
•
The algorithm
The
algorithm ENUMERATE-JOINT-ASK gives inference by enumerating on full joint
distribution.
Characteristics
of algorithm:
1)
It takes input a full joint distribution P and looks up values in it. [The same
algorithm can be modified to take input as Bayesian network and looking up in
joint entries by multiplying the corresponding conditional probability table
entries from Bayesian network.
2)
The ENUMERATION-JOINT-ASK uses ENUMERATION-ASK (EA) algorithm which evaluate
expression using depth-first recursions. Therefore, the space complexity of EA
is only linear in the number of variables. The algorithm sums over the full
joint distribution without ever constructing it explicitely. The time complexity
for network with 'n' boolean variables is always O(2n) which is
better than the O(n 2n) required in simple inferencing approach
(using posterior probability).
3)
The drawback of the algorithm is, it keeps on evaluating repeated sub expression
which results in wastage of computation time.
The
enumeration algorithm for answering queries on Bayesian network.
•
The algorithm
Function
ENUMERATION-ASK (X, e, bn) returns a distribution over X.
Inputs: X, the query variable
e,
observed values for variables E.
bn,
a Bayes net with variables {X} UEU Y/* y = Hidden variable */
Q(X)
← A distribution over X, initially empty for each value xi of X do
extend e with value xi for X.
Q(xi)
← ENUMERATE-ALL (VARS[bn] e) return NORMALIZE (Q(x)).
Function
ENUMERATE-ALL (vars, e) returns a real number.
if
EMPTY? (vars) then return 1.0
Y ←
FIRST (vars)
If Y
has value y in e.
Then
return P(y | parents (Y)) X ENUMERATE-ALL (REST(vars), e)
else
return, Σy Ply parents (Y)) X ENUMERATE-ALL (REST(vars), ey)
where
eyis e extended with Y = y.
Example:
Consider
query,
P(Burglary
| J calls = true, M calls =true)
Hidden
variables in the queries are → Earthquake and Alarm.
using
the query equation.
P(Burglary
| j, m) = α P( Burglary, J, M) = α Σe Σa P(Burglary, e,
a, j, m)
The
semantics of Bayesian networks (equation 7.2.1) then gives us an expression in
terms of CPT entries. For simplicity, we will do this just for Burglary = true.
P(b
| j, m) = α Σe ΣaP(b)P(e)P(a | b, e)P(j | a) P(m | a)
•
To compute this expression, we have to add four terms,
each computed by multiplying five numbers.
•
Worst case, where we have to sum out almost all the
variables, the complexity of the algorithm for a network with n boolean
variables is O(n2n).
•
An improvement can be obtained from the following
simple observations. The P(b) term is a constant and can be moved outside the
summations over a and e, and the P(e) term can be moved outside the summation
over a. Hence, we have
P(b
| j, m) = α P(b) Σe P(e) ΣaP(a | b, e)P(j | a) P(m, a)
This
expression can be evaluated by looping through the variables in order,
multiplying CPT entries as we go. For each summation, we also need to loop over
the variable's possible values. The structure of this computation is shown in
following diagram. Using the numbers from Fig. 7.3.2, we obtain P(b | j, m) = α
× 0.00059224. The corresponding computation for ⌐b yields α × 0.0014919;
Hence,
P(B
| j, m)= α < 0.00059224, 0.0014919 >
≈
<0.284, 0.716 >
That
is, the chance of burglary, given calls from both neighbours is about 28 %. Note
In the Fig. 7.3.7, the evalution proceeds top to down, multiplying values along
each path and summing at the "t" nodes. Observe that there is
repetition of paths for j and m.

3. The Variable Elimination Algorithm
The
enumeration algorithm can be improved substantially by eliminating calculations
of repeated sub expression in tree. Calculation can be done once and save the
results for later use. This is a form of dynamic programming.
Working
of variable elimination algorithm
1)
It works by evaluating expressions such as [P(b | j, m) = α P(b) Σe
P(e) Σa P(a | b, e) P(j | a) P(m | a)] in right-to-left order.
2)
Intermediate results are stored and summations over each variable are done only
for those portions of the expression that depends on the variable.
For
example: Consider the Burglary network.
We
evaluate the expression:
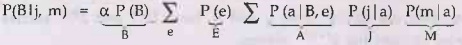
3)
Factors: Each part of the expression is annotated with the name of the
associated variable, these parts are called factors.
Steps
in algorithm:
i)
The factor for M, P (m | a), does not require summing over M. Probability is
stored, given each value of a, in a two-element factor,
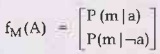
Note fM means that M was used to produce f.
ii)
Store the factor for J as the two-element vector fj (A).
iii)
The factor for A is P(a | B, e) which will be a 2×2×2 matrix fA (A,
B, E).
iv)
Summing out A from the product of these tree factors. This will give 2×2 matrix
whose indices range over just B and E. We put bar over A in the name of the
matrix to indicate that A has been summed out.
FȂJM(B,
E) =Σa fA (a, B, E) × fj (a) × fM
(a)
= fA
(a, B, E) x fJ(a) x fM (a) + fA ( ⌐a, B, E) x
fj (¬a)×fm (¬a)
The
multiplication process used is called a printwise product.
v)
Processing E in the same way (i.e.) sum out E from the product of
fE
(E) and fȂJM (B, E):
fȂJM
(B) = fE (e) × fȂJM (B, e) + fE (¬e) × fȂJM
(B, ¬e).
vi)
Compute the answer simply by multiplying the factor for B. (i.e.) (fB|B)
= P(B), 701 by the accumulated matrix (B) :
P(B
| j, m) = α fB (B) x f EȂJM
(B).
From
the above sequence of steps it can be noticed that two computational operations
are required.
a)
Pointwise product of a pair of factors.
b)
Summing out a variable from a product of factors.
a)
Pointwise product of a pair of factors: The
pointwise product of two factors f1 and f2 yields a new
factor f, those variables are the union of the variables in f1 and f2.
Suppose the two factors have variables Y1,..., Yk. Then
we have f(x1,..., Xj, Y1,.... Yk, Z1,.....Zl)
= f1 (X1,....Xj, Y1.... Yk)
f2(Y1,..... Yk, Z1,....Zl).
If all the variables are binary, then f1 and f2 have 2j+k
and 2k+l entries and the pointwise product has 2j+k+1
entries.
For
example: Given two factors f1 (A, B) and f2 (B, C) with
probability distributions shown below, the pointwise product f1 × f2
is given as f1 (A, B, C).
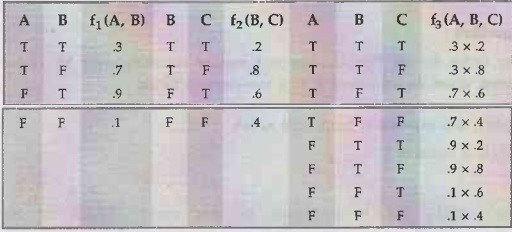
b)
Summing out a variable from a product of factors: It is a straight forward computation. Any factor that does not
depend on the variable to be summed out can be moved outside the summation
process.
For
example:
Σe
fE (e) × fA (A, B, e) × fj (A) × fM
(A) = fj (A) × fM (A) × Σ e fE (e) × fA
(A, B, e). Now, the pointwise product inside the summation is computed and the
variable is summed out of the resulting matrix.
Fj
(A) × fM (A) × Σe fE (e) × fA (A,
B, e) = fj (A) × fM (A) × fEA(A, B).
Matrices
are not multiplied until we need to sum but a variable from the accumulated
product. At that point, multiply those matrices that include the variable to be
summed out.
The
procedure for pointwise product and summing is given below, the variable
elimination algorithm is shown below:
Function ELIMINATION-ASK (X, e, bn) returns a distribution over X
Inputs: X, the query variable
e, evidence specified as an event
bn, a Bayesian network specifying joint distribution P(X1,...,
Xn).
Factors ← [ ] : vars ← REVERSE (VARS [bn])
for each var in vars do
Factors ←[MAKE-FACTOR (var, e) factors]
if var is a hidden variable then
Factors←SUM-OUT (var, factors)
returns NORMALIZE (POINTWISE-PRODUCT (factors)).
P (J
calls, Burglary = True)
The first
step is to write out the nested summation.
P(J
| b) = α P(b) Σe P(e) Σa P(a | b, e) P(J | a) Σm
P(M | a).
Evaluating
this expression from right to left,
Σm
P(M | a) is equal to 1.
Note The variable M is irrelevant to this query. Result of the query P
(J calls/Burglary = True) is unchanged if we remove M calls from the network.
We can remove any leaf node which is not a query variable or an evidence
variable. After its removal, there may be more leaf nodes and they may be
irrelevant. Eventually we find that every variable that is not an ancestor of a
query variable or evidence variable is, irrelevant to the query. A variable
elimination algorithm can remove all these variables before evaluating the
query.
4. The Complexity Involved in Exact Inferencing
The
variable elimination algorithm is more efficient than enumeration algorithm
because it avoids repeated computations as well as drops irrelevant variables.
The
variable elimination algorithm constructs the factor, deriving its operation.
The space and time complexity of variable elimination is directly dependant on
size of the largest factor constructed during the operation. Basically the
factor construction is determined by the order of elimination of variables and
by the structure of the network; which affects both space and time complexity.
For
developing more efficient process we can construct singly connected networks
which are also called as polytrees. In singly connected network, there is at
most one undirected path between any two nodes in the networks. The singly
connected networks have property that, the time and space complexity of exact
inference in polytrees is linear in the size of the network. Here the size is
defined as the number of CPT entries. If the number of parents of each node is
bounded by a constant, then the complexity I will also be linear in the number
of nodes.
For
example: The Burglary network shown in the Fig. 7.3.2 is a polytrees.
[Note
that every problem may not be represented as polytrees].
In
multiply networks [In this, their can be multiple undirected paths between any
two nodes and more than one directed path between some pair of nodes], variable
elimination takes exponential time and space complexity in the worst case, even
when the number of parents per node is bounded. It should be noted that
variable elimination includes inference in propositional logic as a special
case and inference in Bayesian network is NP-hard. In fact it is strictly
harder than NP-complete problem.
Clustering
algorithm:
1)
Clustering algorithm (known as joint tree algorithms) in which inferencing time
can be reduced to O(n). In clustering individual nodes of the network are joint
to form cluster nodes to such a way that the resulting network is a polytree.
2)
The variable elimination algorithm is efficient algorithm for answering
individual queries. Posterior probabilities are computed for all the variables
in the network. It can be less efficient, in polytree network because it needs
to issue O(n) queries costing O(n) each, for a total of O(n2) time,
clustering algorithm, improves over it.

For
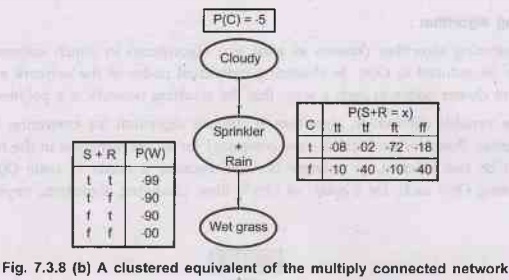
example: The multiply connected network shown in Fig. 7.3.8 (a) can be
converted into a polytree by combining the Sprinkler and Rain node into a
clusternode called Sprinkler + Rain, as shown in Fig. 7.2.8 (b). The two
Boolean nodes are replaced by a meganode that takes on four possible values:
TT, TF, FT, FF. The meganode has only one parent, the Boolean variable. Cloudy,
so there are two conditioning cases.
Peculiarities of algorithm:
1)
Once the network is in polytree form, a special-purpose inference algorithm is
applied. Essentially, the algorithm is a form of constraint propagation where
the constraints ensure that neighbouring clusters agree on the posterior
probability of any variables that they have in common.
2)
With careful book keeping, this algorithm is able to compute posterior
probabilities for all the non-evidence nodes in the network in time O(n), where
n is now the size of the modified network.
3)
However, the NP-hardness of the problem continues: if a network requires
exponential time and space with variable elimination, then the CPTs in the
clustered network will require exponential time and space to construct.
Approximate Inference in Bayesian Network
Because
of the fact that exact inference in multiply connected network is intractable
problem, we go for approximate inferencing.
For
approximate inferencing randomize sampling algorithm (Monte Carlo Algorithm) is
used. Its accuracy depends on number of samples generated. Monte Carlo
Algorithm is widely used in problem areas where it is difficult to calculate
exact quantities.
There
are two families of Monte Carlo algorithm:
1)
Direct sampling algorithm.
2)
Markov chain sampling algorithm.
1. Direct Sampling Algorithm
1)
The basic element in any sampling algorithm is the generation of samples from a
known probability distribution.
For
example: An unbiased coin can be thought of as a random variable coin with
values (heads, tails) and a prior distribution P(coin) <0.5, 0.5>.
Sampling from this distribution is exactly like flipping the coin: with
probability 0.5 it will return 9879 heads, and with probability 0.5 it will
return tails. Given a source of random numbers in the range [0, 1], it is a
simple matter to sample any distribution on a single variable.
2)
The simplest kind of random sampling process for Bayesian networks generates
events from a network that has no evidence associated with it. The idea is to
sample each variable in turn, in topological order.
3)
The probability distribution from which the value is sampled is conditioned on
the values already assigned to the variable's parents.
4)
The sampling algorithm:
Function PRIOR-SAMPLE (bn) returns an event sampled from the prior
specified by bn.
inputs: bn, a Bayesian network specifying joint distribution P(X1,...,
Xn).
X ← an event with n elements.
for i = 1 to n do
xi ← a random sample from P(Xi parent (Xi))
return x.
5)
Applying operations of algorithm on the network in Fig. 7.3.8 (a) assuming an
ordering [Cloudy, Sprinkler, Rain, WetGrass]:
i)
Sample from P(Cloudy) = <0.5, 0.5>; suppose this returns true.
ii)
Sample from P(Sprinkler | Cloudy = true) = <0.1, 0.9>; suppose this
returns false.
iii)
Sample from P(Rain | Cloudy = true) = <0.8, 0.2>; suppose this returns
true. iv) Sample from P(WetGrass | Sprinkler = false, Rain = true) <0.9,
0.1>; suppose this returns true.
In
this case PRIOR-SAMPLE returns the event [true, false, true, true].
6)
PRIOR-SAMPLE generates samples from the prior joint distribution specified by
the nework. First, let Sps (x1,...,xn) be the
probability that a specific event is generated by the PRIOR-SAMPLE algorithm.
Just looking at the sampling process, we have,
Sps
(x1,...,xn)= Πni=1 P(xi
| parents(Xi)).
Because
each sampling step depends only on the parent values. This expression is also
the probability of the event according to the Bayesian net's representation of
the joint distribution. We have,
Sps
(x1,...,xn) = P(x1,...,xn)
7)
In any sampling algorithm, the answers are computed by counting the actual
samples generated. Suppose there are N total samples, and let N (x1,...,xn)
be the frequency of the specific event x1,...,xn. We
expect this frequency to converge in the limit, to its expected value according
to the sampling probability:-
N →
lim ∞ Nps (x1, …., xn )/N = Sps
(x1, …, xn) = P(x1,….xn) ... (7.3.4)
For
example: Consider the event produced earlier : [true, false, true, true]. The
b. a sampling probability for this event is,
Sps
(true, false, true, true) = 0.5 × 0.9 × 0.8 × 0.9
=
0.324
Hence,
in the limit of large N, we expect 32.4 % of the samples to be of this event.
8)
Whenever we use an approximate equality ("≈"), we mean it in exactly
this sense that the estimated probability becomes exact in the large sample
limit. Such an estimate is called consistent.
For
example: One can produce a consistent estimate of the probability of any
partially specified event x1,...,xn, where m≤n, as
follows: -
P(x1,...,xm)
≈ Nps (x1,...,xm)/N ….(7.3.5)
That
is, the probability of the event can be estimated as the fraction of all
complete events generated by the sampling process that match the partially
specified event.
For
example: If we generate 1000 samples from the sprinkler network, and 511 of
them have, Rain = true, then the estimated probability of rain, written as P(Rain
= true), is 0.511.
2. Rejection Sampling in Bayesian Networks
1)
Rejection sampling is a method for producing samples from a hard-to-sample
distribution given an easy-to-simple distribution.
2)
It can be used to compute conditional probabilities; that is, to determine P(X
| e).
3)
The Rejection Sampling algorithm is,
function REJECTION-SAMPLING (x, e, bn, N) returns an estimate of
P(X | e)
inputs: X, the query variable
e, evidence specified as an event.
bn, a Bayesian network.
N, the total number of samples to be generated.
local variables: N, a vector of counts over X, initially zero.
for j = 1 to N do
X ←PRIOR-SAMPLE (bn)
if X is consistent with e is N[x] ← N[x] +1 where x is the value
of X in x.
return NORMALIZE (N[x]).
The
rejection sampling algorithm for answering queries given evidence in a Bayesian
network.
•
Working of algorithm:
i)
It generates samples from the prior distribution specified by the network.
ii)
It rejects all those samples that do not match the evidence.
iii)
Finally, the estimate P(X = x | e) is obtained by counting how often X = x
occurs in the remaining samples.
4)
Let P(X | e) be the estimated distribution that the algorithm returns. From the
definition of the algorithm, we have,
P(X
| e) = α Nps (X, e) = Nps (X, e)/Nps (e)
from
equation (7.3.5) this becomes,
P(X
| e) ≈ P(x, e)/P(e) = P(x, e).
That
is, rejection sampling produces a consistent estimate of the rule probability.
5) Applying operations of algorithm on the network in the Fig. 7.3.8 (a), let
us assume that we wish to estimate P (Rain/Sprinkler = true), using 100
samples. Of the 100 that we generate, suppose 8 have Rain = true and 19 have
Rain = false. Hence,
P(Rain
| Sprinkler = true) ≈NORMALIZE
(<8.19>)
= <0.296, 0.704 >
The
true answer is <0.3, 0.7>
6)
As more samples are collected, the estimate will converges to the true answer.
The standard deviation of the error in each probability will be proportional to
1/√n, where 'n' is the number of samples used in the estimate.
7)
The biggest problem with rejection sampling is that it rejects so many samples
! The fraction of samples consistent with the evidence 'e' drops exponentially
as the number of evidence variables grows, so the procedure is simply unusable
for complex problems.
3. Likelihood Weighing in Bayesian Network
1)
Likelihood weighing avoids the inefficiency of rejection sampling by generating
only events that are consistent with the evidence 'e'.
2)
The Likelihood Weighing algorithm is,
Function
LIKELIHOOD-WEIGHING (X, e, bn, N) returns an estimate of P(X | e) inputs: X,
the query variable
e,
evidence specified as an event
bn,
a Bayesian network.
N,
the total number of samples to be generated.
Local
variables: W, a vector of weighted counts over X, initially zero.
for
j = 1 to N do
X,
w← WEIGHTED-SAMPLE (bn)
W[X]←
W[x] + w where x is the value of X in x.
return
NORMALIZE (W[X]).
Function
WEIGHTED-SAMPLE (bn, e) returns an event and a weight
X ←
an event with n elements; w← 1.
for
i = 1 to n do
if
X, has a value xi in e
then
w ← wX P(Xi = xi parents(Xi))
else
xi ← a random sample from P(Xi | parents (Xi))
return
X, w.
Note
on likelihood weighing algorithm -
i) It
fixes the values for the evidence variables E and samples only the remaining
variables X and Y. This guarantees that each event generated is consistent with
the evidence.
ii) Not
all events are equal, however, before tallying the counts in the distribution
for the query variable, each event is weighted by the likelihood that the event
19 accords to the evidence, as measured by the product of the conditional of
probabilities for each evidence variable, given its parents.
iii)
Intuitively, events in which the actual evidence appears unlikely should be
given less weight.
3)
Applying operations of algorithm on the network Fig. 7.3.8 (a), with the query
P(Rain/Sprinkler = true, WetGrass = true). The process goes as follows:
i)
The weight w is set to 1.0
ii)
Now, an event is generated.
•
Sample from P(Cloudy) = <0.5, 0.5>; suppose
this returns true.
•
Sprinkler is an evidence variable with value true.
Therefore, we set
w ← w P (Sprinkler = true | Cloudy = true) = 0.1.
•
Sample from P(Rain | Cloudy = true) = <0.8,
0.2>; suppose this returns true.
•
WetGrass is an evidence variable with value true.
Therefore, we set
W ← w P(Wet Grass = true | Sprinkler = true, Rain = true) = 0.099.
iii)
Here WEIGHTED-SAMPLE returns the event [true, true, true, true] with weight
0.099, and this is tallied under Rain = true.
iv)
The weight is low because the event describes a cloudy day, which makes the
sprinkler unlikely to be on.
4)
Likelihood Weighting working:
i)Examine
the sampling distribution Sws for WEIGHTED-SAMPLE.
ii)
The evidence variables E are fixed with values 'e'.
iii)
Call the other variables Z, that is, Z = {X}U Y.
iv)
The algorithm samples each variable in Z, in given its parent values:
Sws
(Z, e) =IIli=1 P(Zi | parents (Zi))...
(7.3.6)
Notice
that Parents (Zi) can include both hidden variables and evidence
variables. Unlike the prior distribution P(Z), the distribution Sws
pays some attention to the son evidence the sampled values for each Zi
will be influenced by evidence among Zi's ancestors.
v)
On the other hand, Sws pays less attention to the evidence than does
the true Som posterior distribution P(Z | e), because the sampled values for
each Zi ignore evidence among Zi's non ancestors.
vi)
The likelihood weight w makes up for the difference between the actual and
desired sampling distribution. The weight for a given sample 'x', composed from
Z and 'e', is the product of the likelihood for each evidence variable given
its parents (some or all of which may be among the Zis):
w(Z,
e) = IImj=l P(ei / parents(Ei) .... (7.3.7)
vii)
Multiplying equations (7.3.4) and (7.3.5), we see that the weighted probability
of a sample has the particularly convenient form,
Sws
(Z, e) W(Z, e) = Πli=1 P(yi | parents(Yi))
Πmi=1
P(ei | parents(Ei)) = P(y, e)
Because
the two products cover all the variables in the network, allowing us to use
equation (7.3.1) for the joint probability.
viii)
It can be easy to show that likelihood weighting estimates are consistent. For
any particular value x of X, the estimated posterior probability can be
calculated as follows:
P(x
| e) =α Σy Nws (x, y, e) w (x, y, e)
from
LIKELIHOOD-WEIGHTING.
= α՛ Σy Sws (x, y, e) w (x, y, e)
from
large N.
= α'
Σy Ρ(x, y, e)
= α'
P(x, e) =P(x | e)by equation (7.3.8)
Hence,
likelihood weighting returns consistent estimates.
5)
Performance of algorithm:
i)
Likelihood weighting uses all the samples generated therefore it can be much
more efficient than rejection sampling.
ii)
It will, suffer a degradation in performance as the number of evidence
variables increases.
iii)
Because most samples will have very low weights and hence the weighted estimate
will be dominated by the tiny fraction of samples that accord more than an
infinitesimal likelihood to the evidence.
iv)
The problem is exacerbated if the evidence variables occur late in the variable
ordering, because then the samples will be the simulations that bear little
resemblance to the reality suggested by the evidence.
4. Markov Chain Monte Carlo (MCMC) Algorithm for Inference in Bayesian Networks
Working
of MCMC algorithm
1)
MCMC generates each event by making a random change to the preceding event. It
is therefore helpful to think of the network as being in a particular current
state specifying a value for every variable.
2)
The next state is generated by randomly sampling a value for one of the non-
evidence variables Xi, conditioned on the current values of the
variables in the Markov blanket of Xi.
3)
MCMC therefore wonders randomly around the state space - the space of possible
complete assignments flipping one variable at a time, but keeping the evidence
variables fixed.
4)
For example: Consider the query P(rain | Sprinkler = true, WetGrass = true)
applied to the network of Fig. 7.3.8 (a). The evidence variables 'Sprinkler'
and 'WetGrass' are fixed to their observed values and the hidden variables
'Cloudy'and 'Rain' are initialized randomly. Let us say to true and false
respectively. Thus, the initial state is [true, true, false, true]. Now the
following steps are executed repeatedly:
a)
Cloudy is sampled, given the current values of its Markov blanket variables :
in this case, we sample from P(Cloudy | Sprinkler = true, Rain = false),
(Shortly we will show how to calculate this distribution). Suppose the result
is y = false. Then the new current state is [false, true, false, true].
b)
Rain is sampled, given the current values of its Markov blanket variables : In
this case we sample from P(Rain Cloudy false, Sprinkler = true, WetGrass =
true). Suppose this yields Rain = true. The new current state is [false, true,
true, true].
5)
Each state visited during this process is a sample that contributes to the
estimate, for the query variable Rain. If the process visits 20 states where
Rain is true and 60 states where Rain is false, then the answer to the query is
NORMALIZE (<20, 60>) = <0.25, 0.75>).
6)
The complete algorithm is shown as follows:-
Function MCMC-ASK (X, e, bn, N) returns an estimate of P(X | e)
local variables: N[X], a vector of counts over X, initially zero.
Z, the nonevidence variables in bn.
X, the current state of the network, initially copied from e.
initialize X with random values for the variables in Z.
for j = 1 to N do
N[x] ←N[x] +1 where x is the value of X in x.
for each Zi in Z do...
sample the value of Zi in X from P(Zi |mb(Zi))
given the value of MB (Zi) in X return NORMALIZE (N[X]).
Why
MCMC works
1)
It should be noticed that the sampling process settles into a "dynamic
equilibrium" in which the long-run fraction of time spent in each state is
exactly proporitional to its posterior probability.
2)
This remarkable property follows from the specific transition probability with
which the process moves from one state to another, as defined by the
conditional distribution given the Markov blanket of the variable being
sampled.
3)
Let q(X →X') be the probability that the process makes a transition from state
X to X'. This transition probability defines what is called a Markov chain on
the state space.
4)
Now suppose that we run the Markov chain for 't' steps, and let л(X) be the
probability that the system is in state X at time 't'.
-
Similarly, let лt+1(X') be the probability of being in state X' at
time 't+1.'
-
Given лt (X)
- We
can calculate лt+1(X') by summing for all states the system could be
in at time 't'.
- The
probability of being in that state is the times the probability of making the
transition to X' →πt+1(X') = Σx πt (X) q(X→X')
5)
We will say that the chain has reached its stationary distribution if лt
= πt+1. Call this stationary distribution л; its defining equation
is therefore,
π
(X') = ΣX π (X) q (X→X') for all X' …..(7.3.9)
Under
certain standard assumptions about the transition probability distribution
there is exactly one distribution л satisfying this equation for any given q.
6)
Above equation (7.3.8) can be read as saying that the expected
"outflow" from each state (i.e., its current "population")
is equal to the expected "inflow" from all the state.
One
way to satisfy this relationship is, the expected flow between any pair of
states is the same in both direction. This is the property of detailed balance:
л(X)
q (X→X')=π (X') q (X'→X) for all X, X'. …...(7.3.10)
7)
It can be shown that detailed balance implies stationarily simply by summing
over X in equation (7.3.9). We have,
Σx
π (X) q (X→X') = Σx π (X') q (X' →X) = π (X') Σx q (X' →
X) = π (X').
8)
To show that the transition probability q (X→X') defined by the sampling step
in MCMC-ASK satisfies the detailed balance equation with a stationary
distribution f equal to P(X | e). (The true posterior distribution on the
hidden variables).
This is done in
two steps:-
a)
First, we will define a Markov chain in which each variable is sampled
conditionally on the current values of all the other variables, and we will
show that this satisfies detailed balance.
b)
We will simply observe that, for Bayesian networks, doing that is equivalent to
sampling conditionally on the variable's Markov blanket.
•
Let Xi be the variable to be sampled, and
let ![]() be all the hidden variables other than Xi. Their
values in the current state are xi and
be all the hidden variables other than Xi. Their
values in the current state are xi and ![]() . If we
sample a new xi for Xi conditionally on all the other
variables, including the evidence, we have, q(X→X') = q(xi,
. If we
sample a new xi for Xi conditionally on all the other
variables, including the evidence, we have, q(X→X') = q(xi, ![]() )
→ (xi, xi) = P(xi |
)
→ (xi, xi) = P(xi | ![]() , e).
, e).
•
This transition probability is called Gibbs sampler
and is a particularly convenient form of MCMC. Now we show that the Gibbs
sampler is, in detailed balance with the true posterior.
π
(X) q(X→ X') = P(X | e) P(x'I | ![]() , e) = P(xi,
, e) = P(xi, ![]() | e) P(x'i |
| e) P(x'i | ![]() , e)
, e)
=
P(xi | ![]() , e) P(
, e) P(![]() | e) P(x'i |
| e) P(x'i | ![]() ,
e) (using the chain rule of first term).
,
e) (using the chain rule of first term).
=
P(xi | ![]() , e) P(x'i,
, e) P(x'i, ![]() | e) (using
the chain rule backwards)
| e) (using
the chain rule backwards)
= π
(X') q (X' →X).
•
Note that, a variable is independent of all other
variables given its Markov blanket; hence
P(x'i
+![]() ,e) = P(x' | mb(Xi))
,e) = P(x' | mb(Xi))
where
mb (Xi) denotes the values of the variables in Xi's
Markov blanket, MB (xi).
The
probability of a variable, given its Markov blanket, is proportional to the lo
probability of the variable given its parents times the probability of each
child given its respective parents:-
P(x'
| mb (Xi)) = α P(x' | Parents(Xi)) × II Yj €
children(xi) P(yi | Parents(Yi)) ….(7.3.11)
•
Hence, to flip each variable ![]() , the number of
multiplications required is equal to the number of Xi 's children.
, the number of
multiplications required is equal to the number of Xi 's children.
Artificial Intelligence and Machine Learning: Unit II: Probabilistic Reasoning : Tag: : Probabilistic Reasoning - Artificial Intelligence and Machine Learning - Probabilistic Reasoning and Bayesian Networks
Related Topics
Related Subjects
Artificial Intelligence and Machine Learning
CS3491 4th Semester CSE/ECE Dept | 2021 Regulation | 4th Semester CSE/ECE Dept 2021 Regulation
